|
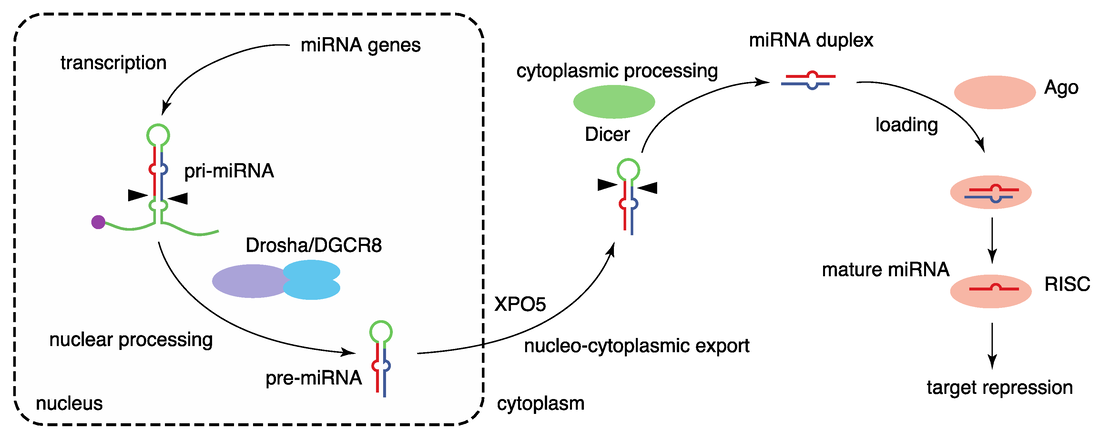
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression post-transcriptionally. The functional mature miRNAs are generated in a multi-stage process and eventually associate with Argonaute proteins to form the core of the miRNA-induced silencing complex (miRISC). miRISC uses the sequence information in the miRNA as a guide to bind complementary sequences on mRNAs. This binding leads to repression of translation and/or mRNA degradation. MiRNAs function in diverse aspects of development and physiology and have been implicated in human disease. In our group, we use various computational techniques to study a wide spectrum of basic and applied questions in miRNA biology.
|
|
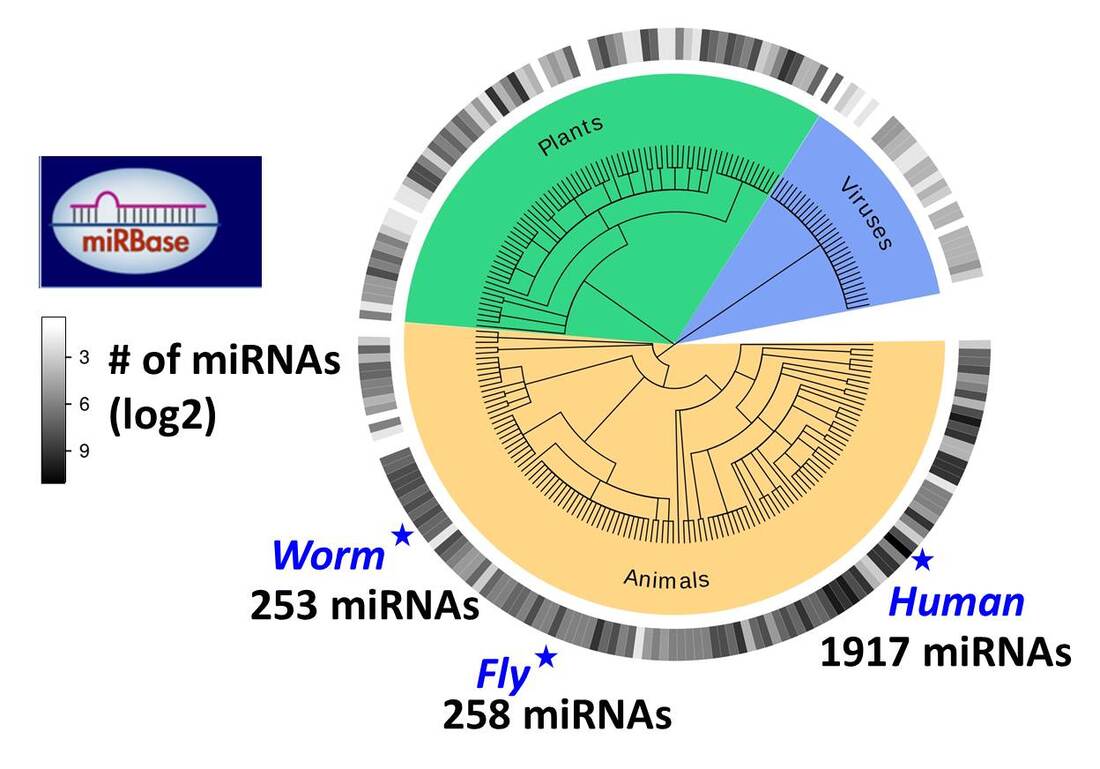
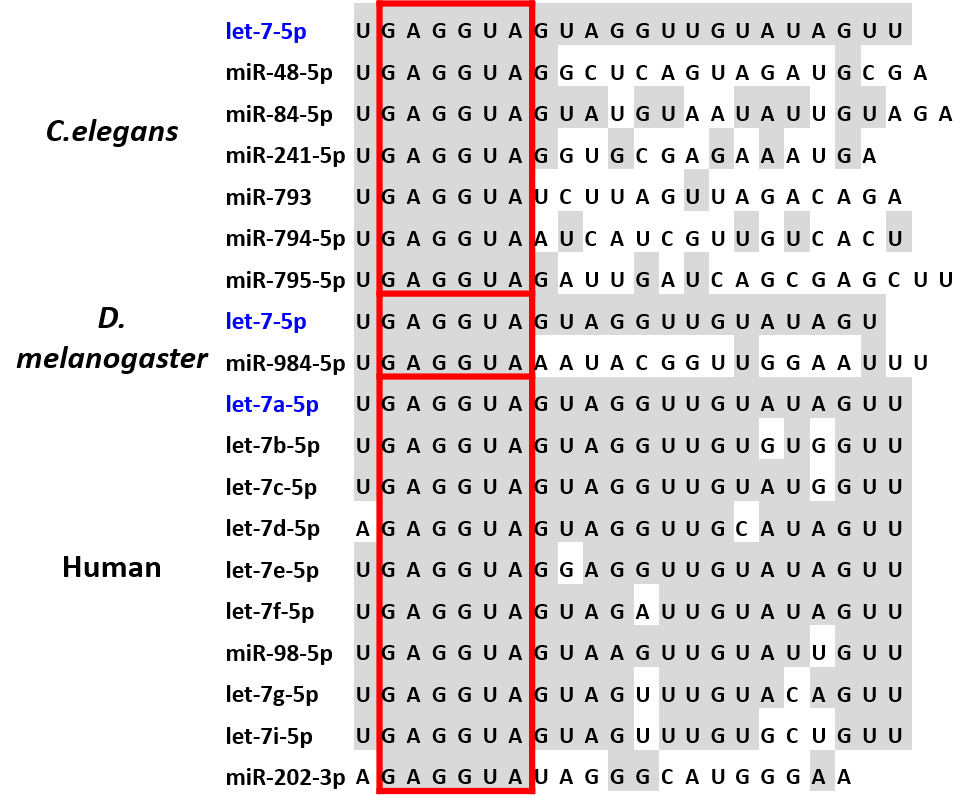
miRNA Evolution
miRNAs are conserved throughout evolution and were shown to be present in all animals and plants and many viruses. We study the evolutionary events that individual miRNA seed families undergo, and the co-evolution between different miRNA families. The evolutionary history of miRNAs can provide useful context for the analysis of their function. |
|
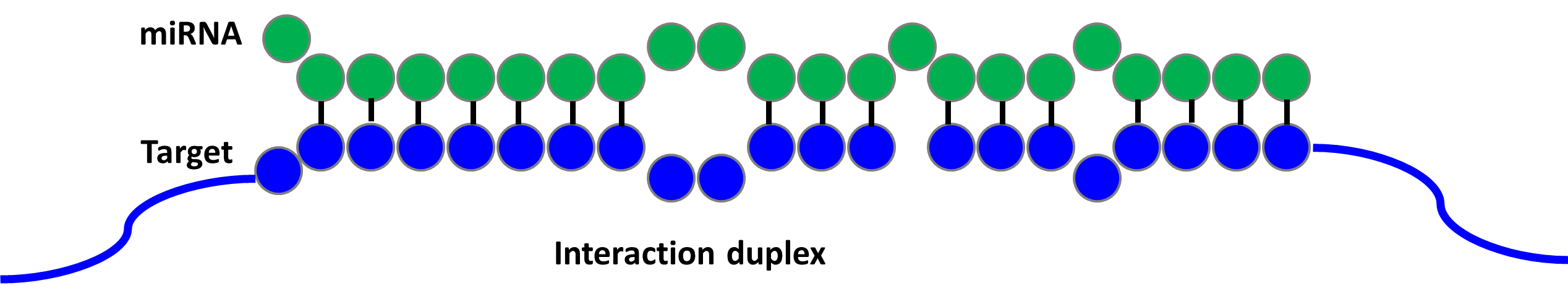
miRNA Target Prediction
The identification of miRNA target sites on mRNAs is a fundamental step in understanding miRNA function. Experimental identification is laborious and time-consuming. We examine experimental datasets of miRNA-target interactions, extract features that describe them, and develop machine learning models to predict new interactions. |
|
miRNAs as biomarkers for human disease
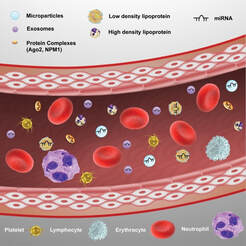
In addition to being expressed inside cells, miRNAs can be detected extracellularly, in body fluids, and hence have emerged as promising non-invasive biomarkers for rapid detection of disease. In collaboration with biologists and clinicians, we evaluate the utility of miRNAs as biomarkers for diagnosis of various pathological conditions. For example, miRNAs as biomarkers for diagnosis and prognosis of acetaminophen liver-toxicity in human patients (PNAS, 2014). |
|
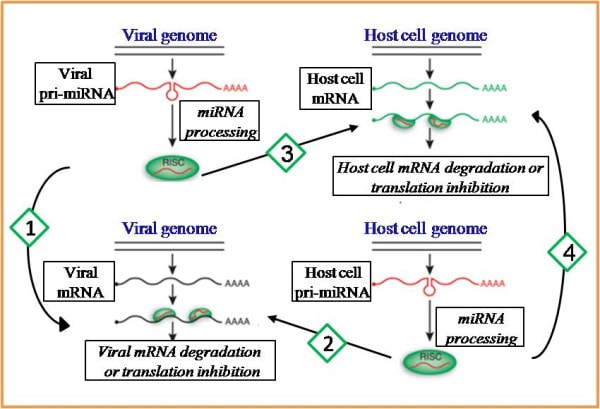
Cross-talk between viruses and their hosts
Some viral organisms also encode miRNAs, contributing to the complex interactions between viruses and their hosts. We developed new bi-clustering algorithms to understand the functional relationship between viral and host miRNAs and their effect on viral and host genes. Specifically, we examined two possible scenarios: (1) Cooperative effects of Epstein Barr Virus (EBV) miRNAs and host miRNAs on the regulation of host genes (BMC Bioinformatics, 2010). (2) Compensating mode of mutual regulation upon Human Cytomegalovirus (HCMV) infection, when host miRNAs are down-regulated and viral miRNAs compensate by mimicking their function (BMC Bioinformatics, 2012). |